
DISGENET plus
Most human diseases are influenced by our genes. Identifying the genes that cause diseases is key to pinpoint novel strategies for prevention and treatment. DISGENET plus is the most comprehensive platform on disease genomics coverage of all therapeutic areas. Up-to-date catalog of genes and genomic variants. Powerful data analytics capabilities. A text mining engine using deep learning approaches, tailored to the peculiarities of the biomedical literature. Developed by the Integrative Biomedical Informatics group, the DISGENET plus API enables programmatic access to the DISGENET plus collections of genes and variants associated with human diseases by means of three different REST endpoints.

ResMarkerDB
Database of Biomarkers of response to monoclonal antibody therapy in breast and colorectal cancer. Developed by the Integrative Bioinformatics Group, it integrates data from existing repositories, and new data extracted and curated from the literature, providing a user-friendly web interface to facilitate the exploration of current knowledge of biomarkers of response in breast and colorectal cancer. ResMarkerDB contains more than 500 biomarker-drug-tumour associations and aims to enhance translational research efforts in identifying actionable biomarkers of drug response in cancer.

GUILDify
GUILDify v2.0 is a Tool to Identify Molecular Networks Underlying Human Diseases. A web server for phenotypic characterization of genes through biological data integration and network-based prioritization algorithms. Towards the goal of extending our knowledge on the genetic elements underlying various phenotypes (including but not limited to disease phenotypes), we aim to use gene-phenotype associations in combination with the network-based prioritization methods. Developed by the Structural Bioinformatics group.

eTOXlab
eTOXlab is a flexible modeling framework. It was developed for supporting models predicting the biological properties of chemical compounds (e.g. QSAR models) in production environments. Developed by the PharmacoInformatics group, in the framework of the eTOX project (GA115002).

ADAN
ADAN (Applicability Domain ANalysis) is a a novel method for assessing the reliability of drug property predictions obtained by in silico methods. The assessment provided by ADAN is based on the comparison of the query compound with the training set, using six diverse similarity criteria. Developed by the PharmacoInformatics group.

SmartAS
SmartAS is a pipeline oriented to finding interesting isoform switches between two conditions, with as many replicates as desired. Developed by the Computational RNA Biology group.
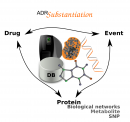
ADRs Substantiation
This is a novel computational framework to aid in the collection and exploration of evidences that support the causal inference of adverse drug reactions (ADRs) detected by mining clinical records. This framework was implemented as publicly available tools integrating state-of-the-art bioinformatics methods for the analysis of drugs, targets, biological processes and clinical events. The availability of such tools for in silico experiments will facilitate research on the mechanisms that underlie ADR, contributing to the development of safer drugs. Developed by the Integrative Bioinformatics group.

ProfileSeq
ProfileSeq is a computational method for the quantitative assessment of biological profiles to provide an exact, nonparametric probability that specific regions of the test profile have higher or lower signal densities than a control set. The method is applicable to high-throughput sequencing data (ChIP-Seq, GRO-Seq, CLIP-Seq, etc.) and to genome-based datasets (motifs, etc.). Developed by the Computational RNA Biology group.

SUPPA
Developed by the Computational RNA Biology group, SUPPA is a tool that generates different Alternative Splicing (AS) events and calculates the PSI ("Percentage Spliced In") value for each event exploiting the fast quantification of transcript abundances from multiple samples.

PsyGeNET
Developed by the Integrative Biomedical Informatics group, PsyGeNET (Psychiatric disorders and Genes association NETwork) is a knowledge platform for the exploratory analysis of psychiatric diseases and their associated genes. PsyGeNET is composed of a database and a web interface supporting data search, visualization, filtering and sharing.



